X marks the spot where ME/CFS biology can be discovered
06 August 2025
Scientists, people with ME/CFS, and their charities came together to create DecodeME, the world's biggest ME/CFS study – and its results are striking. 18,000 people with ME/CFS gave their DNA, enabling DecodeME to reveal eightgenetic signals for the illness. These signals indicate that immune and neurological processes play a significant role in ME/CFS.
Why a DNA study?
We know little about the biology of ME/CFS, which makes it very hard to develop effective treatments. Finding biological differences between people who have ME/CFS and others often doesn’t help because it's usually very hard to tell whether any differences are driving the illness or are simply the result of it. But this isn’t a problem for DNA studies, because diseases don't affect DNA. So genetic differences between those with ME/CFS and those without it must be part of the cause of the disease, not an effect.
DNA carries the instructions for how to make and run a human body. It’s effectively a very long string of four different chemicals, named A, T, C and G for short. These letters spell out our instruction book of life. Some of these letters make up our roughly 20,000 genes, and each gene carries the recipe for making a particular kind of protein. Proteins are our bodies’ ‘doing’ molecules, such as digestive enzymes, antibodies and components of muscle fibres.
Caption: A short section of DNA showing the four chemical letters.
Most proteins do something very specific, so if we can pinpoint which genes are different in people with ME/CFS, we also pinpoint the protein involved – and that points to the biology. By looking at DNA differences, DecodeME was effectively scanning the whole of human biology for problems linked to ME/CFS.
How DecodeME looked for DNA differences
DecodeME was a genome-wide association study (GWAS). These studies don’t look for DNA differences in each of the three billion DNA letters that humans have, because that’s very expensive. Instead, they look only in those places where people often have different letters from each other (each different letter at a particular location is a ‘variant’). DecodeME looked at nearly nine million of the DNA letters, but that is still only three in every thousand letters.
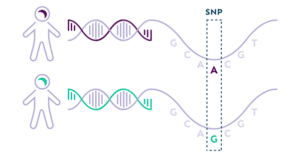
Caption: Illustration of a DNA variant where one person has DNA with the letter A, while another has the letter G at the same position.
DecodeME’s findings
DecodeME looked at the DNA of 15,579 people with an ME/CFS diagnosis who met the study’s tight research criteria and compared it with the DNA of nearly 260,000 controls without ME/CFS. All had European ancestry, which makes the results more robust. A later analysis will look at people of all ancestries.
The analysis revealed eight genetic ‘signals’ for ME/CFS from the nine million variants studied. And these signals will help to reveal the biology of the disease.
You can see these signals as the peaks (‘skyscrapers’) that stick up above the dotted line in the ‘Manhattan plot’ below. They show where there are groups of variants that are significantly different in people with ME/CFS compared to healthy people. These peaks, or signals, are each flagged with the name of the gene that could explain them.
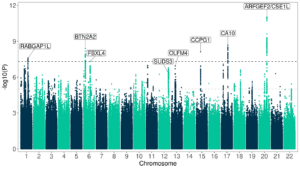
Caption. Manhattan plot for the main DecodeME GWAS.
Six were above the line for DecodeME’s main GWAS. Two others, which were just below the threshold here, were above the threshold when using a smaller group of healthy people for comparison than the main GWAS. And one of those was also significant when restricting the GWAS to the cases who had had an infectious onset.
The genetic signals are like crosses on a treasure map, pointing to hidden biology
GWAS are an affordable way to scan the human genome but are harder to interpret because they only look directly at a few DNA letters in every thousand. This means that although there are eight genetic signals of closely-grouped variants significantly associated with ME/CFS, the DNA defined by the signal will contain many unseen variants. These unseen variants are more likely to be a cause of ME/CFS because there are far more of them.
So each genetic signal is like an ‘X’ on a treasure map indicating roughly where the researchers should dig for treasure. The next step is to find that treasure – the specific genes that are causing ME/CFS.
DecodeME identifies top genes
DecodeME has started searching for the treasure, and has identified likely genes using several methods. These methods looked at genes within the genetic signals – the peaks on the Manhattan plot.
The most powerful method searched those genes to find ones whose activity levels (the amount of protein they make) are known to go up or down depending on what form a particular variant within the ME/CFS genetic signal takes. This means that the activity of those genes is related to someone having ME/CFS – a strong clue that those genes might be a cause of ME/CFS.
DecodeME identified 29 top gene candidates this way, and many of them are immune genes. The Manhattan plot shows the (typically unpronounceable) names of the eight strongest candidates. One of them, called RABGAP1L, produces a protein that helps fight off some viruses and bacteria – and DecodeME’s analysis showed that having ME/CFS is associated with having less of this protective protein.
Another gene on the plot, SUDS3, produces a protein that dampens down the inflammatory response of the brain’s main immune cells, called microglia. So people with ME/CFS could have a weaker microglial inflammatory response in the brain.
A third example from the plot is a gene called CA10, which has been strongly linked with chronic pain – a major feature of ME/CFS.
DecodeME used another analysis method called MAGMA to link ME/CFS genetic signals to the nervous system. MAGMA first identified a set of 13 genes associated with ME/CFS signals. Then, using existing data on gene activity, it found that this set of genes was more active than expected in 13 types of tissue in the human body (out of a possible 54), and all 13 tissues were brain regions.
DecodeME also looked at genetic signals associated with depression and anxiety that were near the genetic signals found for ME/CFS, and found that neither involved the same causal variants as ME/CFS.
So, DecodeME showed that immunological and neurological processes are likely to play a part in ME/CFS. They also found a link to chronic pain, but not to anxiety or depression.
What next?
DecodeME aims to do more analysis to refine and add to these findings. This includes analysing the sex chromosomes, which might show why far more women than men get ME/CFS (DecodeME’s genetic results to date do not). They will also look more deeply at HLA genes, an important group of immune genes that often affect the risk of getting diseases, but for which DecodeME’s initial results were not clear-cut.
But the real story here is that DecodeME has generated a host of new biological clues.
Researchers could flood in to transform the field
Professor Chris Ponting, the lead scientist on DecodeME, stressed that GWAS findings are robust and should provide a firm foundation for future research. He said, ’We need researchers whose expertise is directly relevant to these eight genetic signals - especially those who've never worked on ME/CFS before - to come forward and help us explain more precisely what DecodeME's signals mean.’
Sonya Chowdhury, chair of DecodeME’s management group and CEO of Action for ME, said, ‘These results are groundbreaking. With DecodeME, we have gone from knowing next to nothing about the causes of ME/CFS, to giving researchers clear targets.’
She added, ‘We hope this attracts researchers, drug developers, and fair funding to ME/CFS – and speeds up the discovery of treatments.’