What we learned from the current largest genetic study into M.E./CFS
DecodeME will be the world’s largest genetic study of Myalgic encephalitis (ME) / Chronic Fatigue Syndrome (CFS). The current largest is by the UK Biobank, which analysed the DNA of people with self-reported CFS. Has the study help unravel the genetic secrets of ME/CFS? Here’s what we learned from a paper just published.
Quick summary: The study did not find a gene with rare DNA changes which make people with those changes more likely to develop ME/CFS. Why was a genetic signal not found? Likely reasons include: many more people need to be studied, or a narrower definition of ME/CFS is needed.
The background: The UK Biobank undertook a genetic analysis of 1,232 people with self-reported CFS, alongside thousands of other medical conditions.
DecodeME uses a method called a genome-wide association study (GWAS) to focus on DNA letters that are often different among humans. This UK Biobank project focuses, instead, on very rare DNA letters. They wanted to see whether these rare changes often hit the same gene for a particular condition.
A similar approach, using much less data, had previously been used to support the IDO Metabolic Trap Hypothesis that rare changes in a gene called IDO2 predispose people to ME/CFS. Using the new data we can immediately see whether any gene is often damaged in ME or ME/CFS.
The results: If IDO2, or any gene, has many more rare damaging DNA changes in people with CFS (compared with others) then its data point – the coloured triangles – would be higher up on the graph than data points for genes whose DNA doesn’t affect CFS. To be significant a gene’s data point should lie above one or both of two thresholds, shown in the plot as horizontal blue dashed lines.
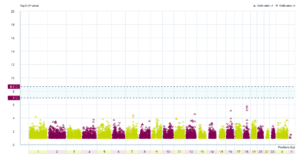
Graph showing genetic changes across genes
As you can see, there is no gene (not even IDO2) that has suffered many more rare damaging changes in people with CFS than in others. This is disappointing. If there was such a gene then scientists could immediately begin to experimentally investigate it.
So why was there no “genetic signal”, despite this large study of 1,232 people? A likely reason is that the DNA of many, many more people is needed to detect this signal – the study is “under-powered”. Another possible reason is that people in this study had self-reported being diagnosed with CFS, and a narrower definition of ME (or ME/CFS) would be needed to improve the signal.
How does this relate to the DecodeME study?
DecodeME is focusing first on DNA letters that are often different between people, and not on rare DNA changes, as in this UK Biobank analysis. This is because genome sequencing technology – needed to find rare DNA changes – is about ten-times more expensive than surveying common DNA differences.
However, DecodeME does plan to store a DNA sample from anyone who provides their consent. This means that – as soon as possible, once additional funds are secured – rare DNA variants from many more thousands of people with ME/CFS can be sequenced.
UK Biobank’s research announcement reveals the scale of the challenge to find genetic signals of ME/CFS. It is clear that there is no “ME/CFS gene”, or even a handful of such genes. Instead, each person with a genetic predisposition to ME/CFS will have many DNA changes – both rare and common – that have tipped the balance in favour of disease over health.
It is our goal to begin cataloguing these DNA changes. This will eventually help scientists work out how to intervene and tip the balance back towards health.
Join us or find out more:
Want to help unravel the genetic secrets of ME/CFS? Register your interest in taking part or hearing more.
Want to know more about the science behind our study? Read more.